Abstract
Introduction: Whole exome sequencing (WES) has identified somatic mutations significant to the pathogenesis of MDS, in which germline tissue sequencing is a challenging, yet vital, component of genetic discovery. Challenges include deriving germline DNA of sufficient quantity and quality that is devoid of neoplastic contamination. To identify the optimal control for The National MDS Natural History Study conducted by the NHLBI and the NCI, we perform a comprehensive assessment of an array of candidate germline tissues. This study will provide a future resource for genomic research in MDS and related hematopoietic diseases.
Methods: Twenty-six MDS cases were included and up to six candidate germline tissues were collected for each case: ten eyebrow hair follicles/patient (n=26), a 2-4 mm skin biopsy (n=26), 50 ml urine (n=26), ten fingernail clippings/patient (n=3), purified CD3+ T-cells from peripheral blood (n=26), and buccal swabs (n=11). In addition, bone marrow mononuclear cells (BMNCs) were collected and analyzed from each case. DNA quantity, by Qubit quantification, and DNA quality, by an Illumina TapeStation, were measured on all samples. WES was performed with SureSelect (Agilent) exome enrichment on native and whole genome amplified (WGA) DNA (Illustra GenomiPhi V2 kit) from BMNCs and sequenced on a HiSeq instrument (Illumina). WGA was performed in germline tissue with less than 1.5µg total DNA (eyebrow hair and skin). Sequence reads were aligned with the Burrows-Wheeler Aligner and variant calling performed by Strelka/MuTect. Targeted resequencing using NimbleGen Hybrid Capture System (Roche) was used to confirm nucleotide variants detected by Strelka/MuTect software using native (non-WGA) DNA. False-negative and false-positive variants were defined as those missed in 1 of 4 or those identified in 1 of 4 BMNC/germline pairs, respectively.
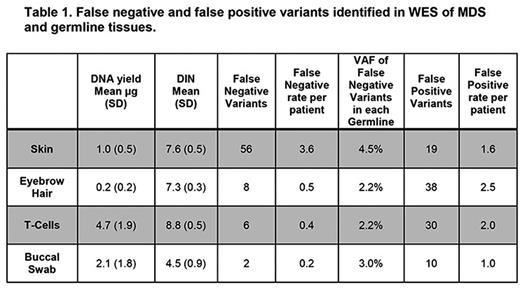
Results: All patients met a WHO-defined MDS diagnosis and had active disease at the time of sampling. Sequencing was performed on DNA from four candidate germline controls (skin, hair, ≥ 95% pure CD3+ T-cells, and buccal swabs as summarized in Table 1) from 16 MDS cases, and on DNA from the BMNCs. Buccal cells were analyzed on 11 of the cases. Urine and fingernail clippings were excluded from WES due to morphologic leukocyte contamination and poor DNA quality (mean DIN=1.6, n=3), respectively. The median coverage was 92x for WES and 404x for targeted resequencing.
Strelka/MuTect identified 272 protein-coding variants in at least one BMNC-germline pair including 28 (10.2%) known variants (Papaemmanuil et al, Blood . 122:3616-3627, 2013). Of 272 total variants, 133 variants (48.8%, 8.3 variants/patient) were identified in all BMNC-germline pairs. However, a high rate of false-negative variants was seen when skin was used as the control (20%, 56 total, 3.5 variant/patient). False positive and false negative variants are summarized in Table 1.
To explore missed variant calls, we compared the coverage and variant allele frequency (VAF) for all variants in the BMNCs and germline controls. No significant differences were seen in BMNC coverage, BMNC VAF, or germline tissue coverage at false negative variants (Table 1). However, increases were identified in the VAF of false negative variants in the skin (29 variants, mean VAF 5.8%, range 0-8.5%) suggesting neoplastic contamination. This result is in agreement with the observation that a larger fraction of variants were missed in BMNC-skin pairs.
Given these results, we tested whether another variant caller using distinct statistical methodology would perform more favorably using skin as a germline control. VarScan employs a heuristic approach hypothesized to overcome neoplastic contamination. VarScan was able to identify variants missed by Strelka/MuTect in a subset of variants (27 of 56) with skin. However 12 of 56 positions were not called by VarScan against any germline control suggesting differences between methods.
Conclusions: Identifying the bioinformatics pipeline and germline tissue that best identifies somatic variants given tumor contamination, while considering the quantity and quality of DNA obtained, may augment the spectrum of mutations seen in MDS. Skin biopsies can be contaminated with neoplastic variants, which can result in missed variant identification. Future sequencing studies should consider this and explore non-skin germline control tissues to map the MDS mutatome.
Padron: Incyte: Honoraria, Research Funding. Teer: Interpares Biomedicine: Consultancy. Komrokji: Novartis: Honoraria, Speakers Bureau; Celgene: Honoraria. Sekeres: Celgene: Membership on an entity's Board of Directors or advisory committees. Bejar: Genoptix: Consultancy, Honoraria, Patents & Royalties; Celgene: Consultancy, Honoraria, Other: DSMB, Steering Committee, Research Funding; Otsuka/Astex: Honoraria, Other: Ad-hoc advisory board; Foundation Medicine: Honoraria, Other: Ad-hoc advisory board; AbbVie/Genetech: Honoraria, Other: Ad-hoc advisory board; Modus Outcomes: Consultancy, Honoraria. Epling-Burnette: Forma Therapeutics: Research Funding; Incyte: Research Funding; Celgene: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal